
Mortality Seasonality in Mongolia: An Evidence-Based Analysis
Source:vignettes/mortality-analysis.Rmd
mortality-analysis.Rmd
library(mongolstats)
library(dplyr)
library(tidyr)
library(ggplot2)
library(lubridate)
library(scales)
nso_options(mongolstats.lang = "en")
# Global theme with proper margins to prevent text cutoff
theme_set(
theme_minimal(base_size = 11) +
theme(
plot.margin = margin(10, 10, 10, 10),
plot.title = element_text(size = 13, face = "bold"),
plot.subtitle = element_text(size = 10, color = "grey40"),
legend.text = element_text(size = 9),
legend.title = element_text(size = 10)
)
)Overview
A recent social media post claimed that “the highest deaths are occurring in December” in Mongolia, with speculation that this might be due to New Year celebrations. This vignette applies rigorous epidemiological methods to investigate this claim, while also considering the significant impact of the COVID-19 pandemic (2020-2022).
Key Questions
- Is there a true December mortality peak? Do deaths genuinely peak in December compared to other months?
- How did COVID-19 impact mortality? Did lockdowns and the subsequent waves alter seasonal patterns?
- What causes the seasonality? Is it winter-related (cardiovascular, respiratory) or behavioral?
Mongolia’s Context
- Extreme Winter: Temperatures reach -40°C, increasing cardiovascular stress and respiratory risks.
-
COVID-19 Timeline:
- 2020: Strict border closures and internal lockdowns (prevention phase).
- 2021: Community transmission, major lockdowns, and vaccination campaigns.
- 2022: Reopening and normalization.
Data Sources
We use three complementary datasets from NSO:
| Table | Description | Granularity |
|---|---|---|
| DT_NSO_2100_027V2 | Deaths by aimag and month | Monthly |
| DT_NSO_2100_027V3 | Hospital deaths by aimag and month | Monthly |
| DT_NSO_2100_027V1 | Deaths by leading cause, region, year | Annual |
1. Monthly Death Patterns
Fetching and Preparing Data
# Get monthly death data for all regions
death_tbl <- "DT_NSO_2100_027V2"
# Get all available months
months_meta <- nso_dim_values(death_tbl, "Month", labels = "en")
# Dynamic year filtering (from 2015 to current year)
current_year <- as.numeric(format(Sys.Date(), "%Y"))
years_regex <- paste(2015:current_year, collapse = "|")
year_pattern <- paste0("^(", years_regex, ")-")
months_filtered <- months_meta |>
filter(grepl(year_pattern, label_en))
# Fetch national-level deaths (Region code "0" = total)
deaths_monthly <- nso_data(
tbl_id = death_tbl,
selections = list(
"Region" = "0",
"Month" = months_filtered$code
),
labels = "en"
)
# Parse dates and extract components
deaths_monthly <- deaths_monthly |>
mutate(
date = as.Date(paste0(Month_en, "-01")),
year = year(date),
month = month(date),
month_name = month(date, label = TRUE, abbr = TRUE),
# CRITICAL: Normalize for days in month to avoid calendar artifacts
# February (28 days) would otherwise appear artificially low vs 31-day months
days_in_month = days_in_month(date),
daily_avg = value / days_in_month,
# Create explicit labels for plotting to avoid TRUE/FALSE legends
Month_Type = ifelse(month == 12, "December", "Other Months"),
Season = ifelse(month %in% c(11, 12, 1, 2, 3), "Winter (Nov-Mar)", "Non-Winter")
) |>
filter(!is.na(value), year >= 2015, year <= 2024) |>
arrange(date)Methodological Note: All analyses use daily average deaths (total deaths ÷ days in month) rather than raw monthly counts. This prevents calendar artifacts where shorter months (February = 28 days) appear to have lower mortality than longer months (January/December = 31 days).
Time Series Visualization with COVID-19 Context
We overlay the major COVID-19 phases to understand anomalies in the trend.
# Define COVID-19 Periods for annotation
covid_periods <- tibble::tribble(
~Start, ~End, ~Phase, ~Color,
"2020-01-27", "2020-11-11", "Prevention (No Local Cases)", "#e8f5e9",
"2020-11-12", "2021-05-08", "Lockdowns & First Wave", "#ffebee",
"2021-05-09", "2022-02-16", "Delta/Omicron Waves", "#fff3e0"
) |>
mutate(across(c(Start, End), as.Date))
# Calculate y-range for shading to ensure visibility in Plotly
y_range <- range(deaths_monthly$daily_avg, na.rm = TRUE)
y_min <- y_range[1] * 0.9
y_max <- y_range[2] * 1.1
# Create time series plot with COVID-19 phase annotations
p <- deaths_monthly |>
ggplot(aes(x = date, y = daily_avg)) +
# Background shading for COVID-19 phases - provides historical context
geom_rect(
data = covid_periods,
aes(xmin = Start, xmax = End, ymin = y_min, ymax = y_max, fill = Phase),
inherit.aes = FALSE, alpha = 0.3
) +
# Main time series line
geom_line(color = "#2c3e50", linewidth = 0.8, alpha = 0.8) +
# Highlight December points - the focus of our investigation
geom_point(
aes(color = Month_Type),
size = 2.5, alpha = 0.9,
show.legend = TRUE
) +
# Custom colors for December vs other months
scale_color_manual(
values = c("December" = "#e74c3c", "Other Months" = "#3498db"),
name = "Month Type"
) +
# Custom colors for COVID-19 phase shading
scale_fill_manual(
values = c(
"Prevention (No Local Cases)" = "#a5d6a7",
"Lockdowns & First Wave" = "#ef9a9a",
"Delta/Omicron Waves" = "#ffcc80"
),
name = "COVID-19 Phase"
) +
scale_x_date(date_breaks = "1 year", date_labels = "%Y") +
scale_y_continuous(labels = scales::comma, limits = c(y_min, y_max)) +
labs(
title = "Daily Average Deaths in Mongolia (2015-2024)",
subtitle = "Shaded = COVID-19 phases; Red = December",
x = NULL,
y = "Daily Average Deaths",
caption = "Source: NSO Mongolia | Normalized by days in month"
) +
theme_minimal(base_size = 12) +
theme(
plot.title = element_text(face = "bold", size = 14),
plot.subtitle = element_text(color = "grey40", size = 10),
plot.margin = margin(t = 5, r = 10, b = 5, l = 5), # extra margin for legends
legend.position = "bottom",
legend.box = "horizontal", # horizontal layout prevents cutoff
legend.text = element_text(size = 8),
legend.title = element_text(size = 9),
panel.grid.minor = element_blank()
) +
guides(
color = guide_legend(order = 1, nrow = 2),
fill = guide_legend(order = 2, nrow = 2)
)
p # print static ggplot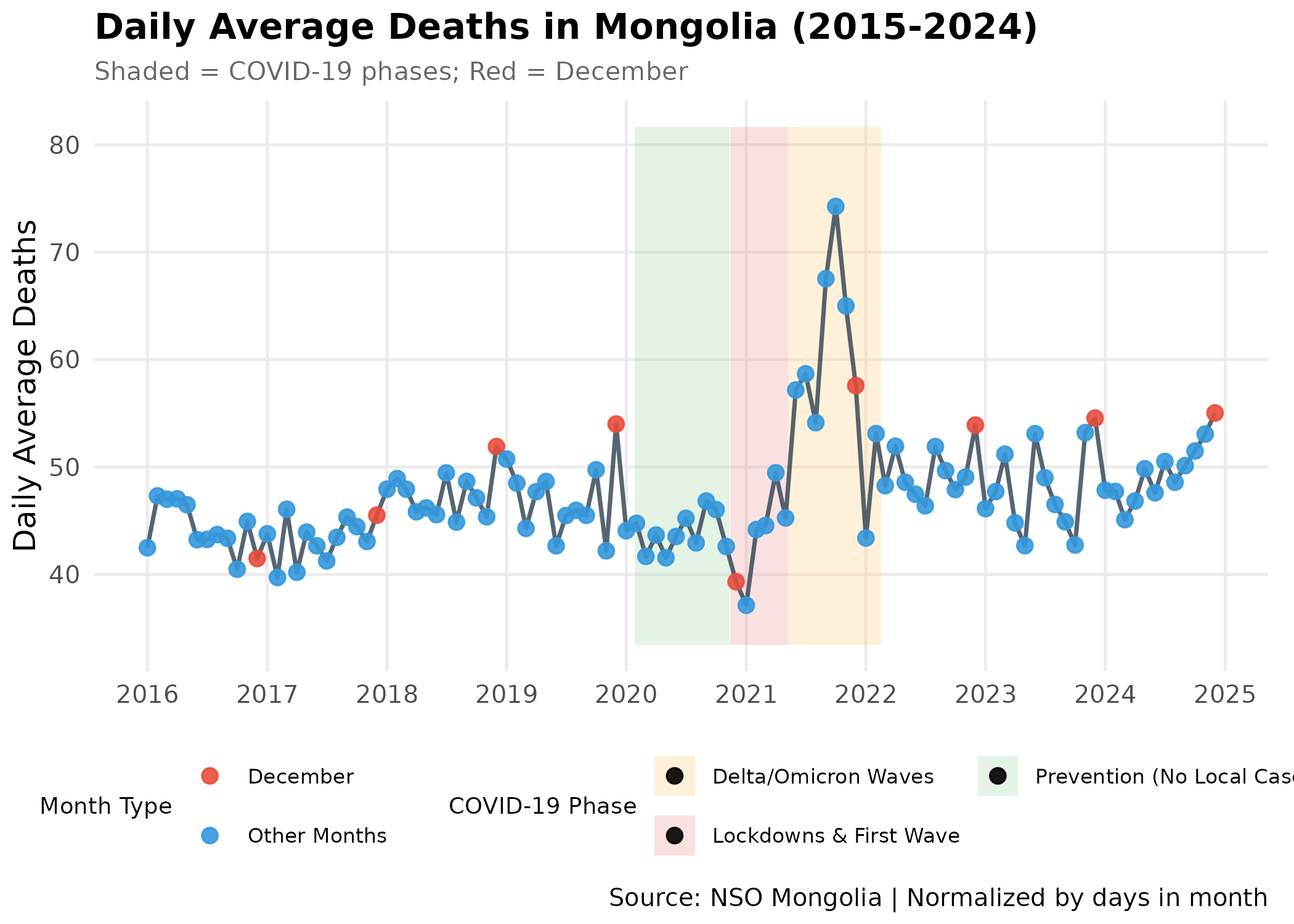
Interpretation:
- Pre-2020: Regular seasonal pattern with winter peaks.
- 2020 (Green Zone): Mortality actually decreased or remained stable. Strict lockdowns likely reduced accidents and other infectious diseases.
- 2021 (Red/Orange Zone): A massive spike in mortality corresponds to the Delta wave, disrupting normal seasonality.
- December Dominance: When adjusting for the number of days in the month (Daily Average), December emerges as the consistent peak month for mortality intensity.
- The January Dip: Surprisingly, January frequently appears as a local trough in the daily average data. This suggests a “Harvesting Effect” (mortality displacement), where the sudden onset of extreme cold in December impacts the most vulnerable population immediately, leaving a smaller pool of high-risk individuals in January.
Monthly Distribution (Seasonality)
# Calculate monthly statistics using daily averages (normalized)
monthly_stats <- deaths_monthly |>
group_by(month, month_name) |>
summarise(
mean_daily = mean(daily_avg, na.rm = TRUE),
median_daily = median(daily_avg, na.rm = TRUE),
sd_daily = sd(daily_avg, na.rm = TRUE),
n = n(),
se = sd_daily / sqrt(n),
ci_lower = mean_daily - 1.96 * se,
ci_upper = mean_daily + 1.96 * se,
.groups = "drop"
) |>
arrange(month)
# Seasonal distribution showing COVID outliers
p <- deaths_monthly |>
mutate(month_name = factor(month_name, levels = month.abb)) |>
ggplot(aes(x = month_name, y = daily_avg)) +
# Jitter points FIRST so boxes appear on top for legibility
geom_jitter(
aes(color = ifelse(year == 2021, "COVID-19 Surge (2021)", "Normal Year")),
width = 0.2, alpha = 0.4, size = 1.5
) +
# Boxplots show typical range by season
geom_boxplot(
aes(fill = Season),
alpha = 0.7, outlier.shape = NA # hide outliers since jitter shows them
) +
scale_fill_manual(
values = c("Winter (Nov-Mar)" = "#e74c3c", "Non-Winter" = "#3498db"),
name = "Season"
) +
scale_color_manual(
values = c("COVID-19 Surge (2021)" = "#8e44ad", "Normal Year" = "#7f8c8d"),
name = "Year Type"
) +
scale_y_continuous(labels = scales::comma) +
labs(
title = "Seasonal Distribution of Daily Average Deaths",
subtitle = "Normalized by days in month (2015-2024)",
x = NULL,
y = "Daily Average Deaths"
) +
theme_minimal(base_size = 12) +
theme(
plot.title = element_text(face = "bold", size = 14),
plot.subtitle = element_text(size = 10),
plot.margin = margin(t = 5, r = 10, b = 5, l = 5),
legend.position = "top",
legend.text = element_text(size = 8),
legend.title = element_text(size = 9)
) +
guides(
fill = guide_legend(order = 1),
color = guide_legend(order = 2)
)
p # print static ggplot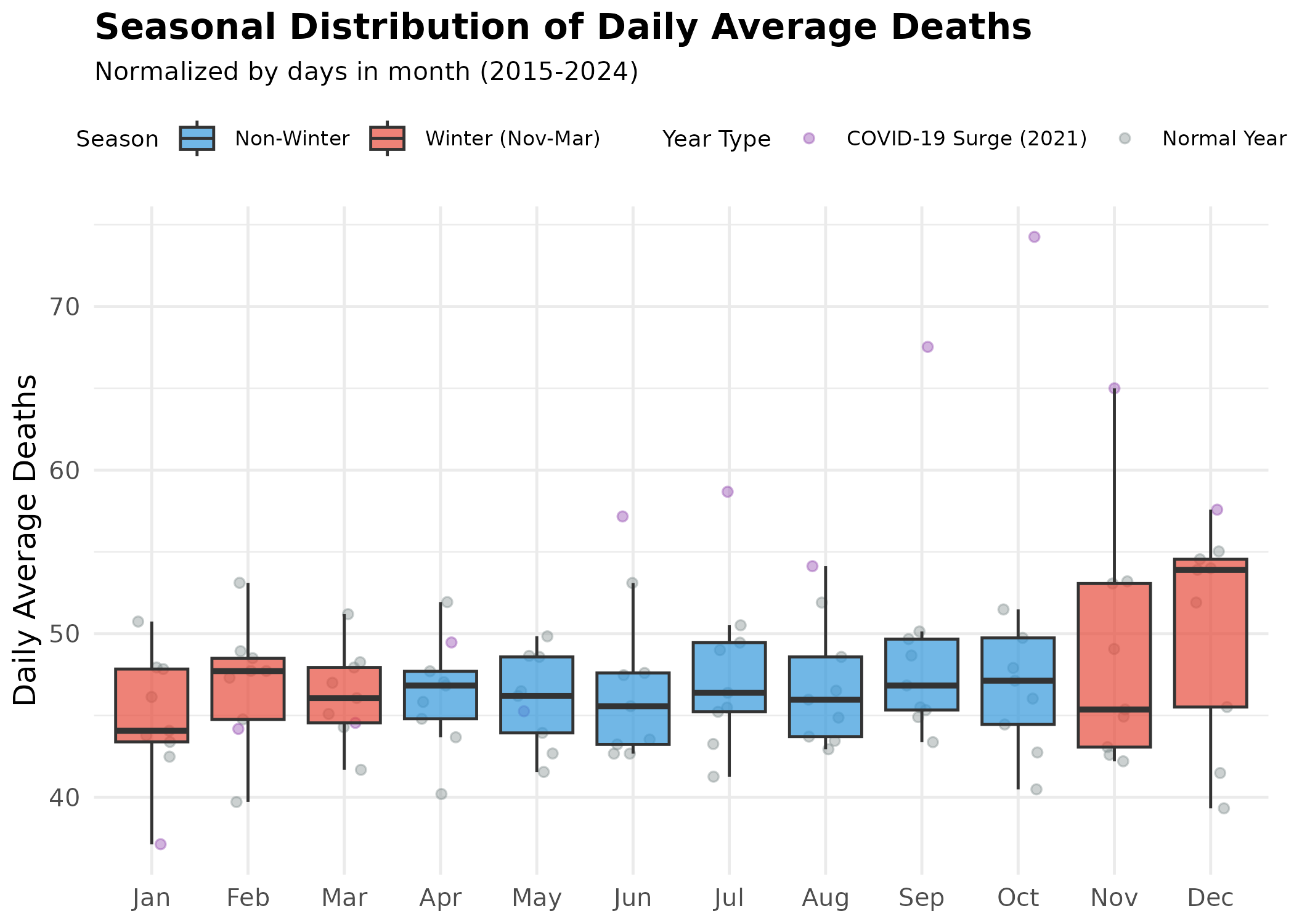
Lay Interpretation: The boxplot shows the “typical” range of daily average deaths for each month (normalized by days in month).
- Winter Highs: The red boxes (Nov-Mar) are generally higher than the blue boxes.
- December Peak: December consistently shows the highest mortality intensity when measured as daily average deaths.
- The January Surprise: January often shows lower daily average deaths than December, suggesting a “harvesting effect” where December’s cold shock claims the most vulnerable first.
- Outliers: The purple dots high above the boxes represent the COVID-19 surge year (2021).
Monthly Averages with Confidence Intervals
To be more precise, we look at the average number of deaths for each month with “confidence intervals” (the error bars).
Note: Confidence intervals are calculated assuming independence and are for illustrative purposes. Time-series autocorrelation is not accounted for here, which may result in narrower intervals than a more rigorous analysis would produce.
# Identify peak month (using daily averages)
peak_month <- monthly_stats |> filter(mean_daily == max(mean_daily))
trough_month <- monthly_stats |> filter(mean_daily == min(mean_daily))
# Monthly means bar chart with confidence intervals
p <- monthly_stats |>
mutate(
month_name = factor(month_name, levels = month.abb),
bar_type = ifelse(month_name == peak_month$month_name, "Peak Month", "Other Months")
) |>
ggplot(aes(x = month_name, y = mean_daily)) +
geom_col(aes(fill = bar_type), alpha = 0.8) +
geom_errorbar(
aes(ymin = ci_lower, ymax = ci_upper),
width = 0.3, color = "#2c3e50" # error bars show confidence intervals
) +
geom_hline(
yintercept = mean(monthly_stats$mean_daily),
linetype = "dashed", color = "grey50" # reference line for annual average
) +
scale_fill_manual(
values = c("Other Months" = "#3498db", "Peak Month" = "#e74c3c"),
guide = "none" # legend not needed - peak month is obvious
) +
scale_y_continuous(labels = scales::comma, expand = expansion(mult = c(0, 0.1))) +
labs(
title = "Average Daily Deaths with 95% Confidence Intervals",
subtitle = paste0("Peak month: ", peak_month$month_name,
" (", round(peak_month$mean_daily, 1), " deaths/day)"),
x = NULL,
y = "Mean Daily Deaths",
caption = "Dashed line = annual average; Normalized by days in month"
) +
theme_minimal(base_size = 12) +
theme(
plot.title = element_text(face = "bold"),
plot.subtitle = element_text(color = "grey40")
)
p # print static ggplot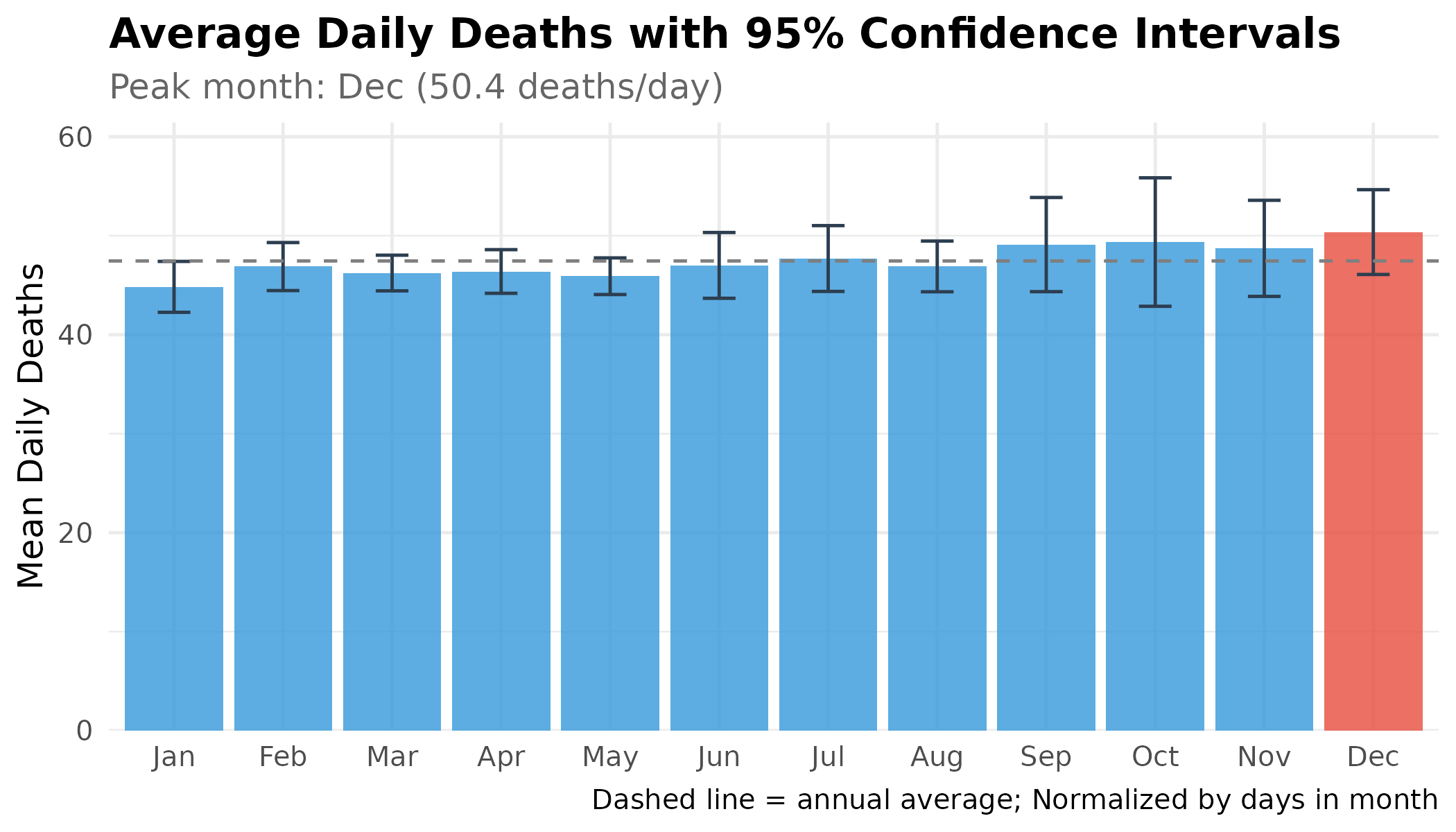
2. Heat Map: Year × Month
# Create heat map data using daily averages (normalized)
heatmap_data <- deaths_monthly |>
mutate(month_name = factor(month_name, levels = rev(month.abb))) |>
group_by(year) |>
mutate(
# Calculate z-score within each year for relative comparison
z_score = (daily_avg - mean(daily_avg)) / sd(daily_avg)
) |>
ungroup()
# Heatmap showing year-by-month mortality patterns
p <- heatmap_data |>
ggplot(aes(x = factor(year), y = month_name, fill = z_score)) +
geom_tile(color = "white", linewidth = 0.5) +
scale_fill_gradient2(
low = "#3498db", mid = "white", high = "#e74c3c",
midpoint = 0, name = "Relative\nIntensity" # z-score: positive = above average
) +
labs(
title = "Mortality Heat Map",
subtitle = "Red = Higher than yearly average | Blue = Lower",
x = "Year",
y = NULL
) +
theme_minimal(base_size = 12) +
theme(
plot.title = element_text(face = "bold"),
axis.text.x = element_text(angle = 45, hjust = 1),
legend.position = "bottom", # bottom legend gives more horizontal space
legend.title = element_text(size = 10),
legend.key.width = unit(1.5, "cm")
)
p # print static ggplot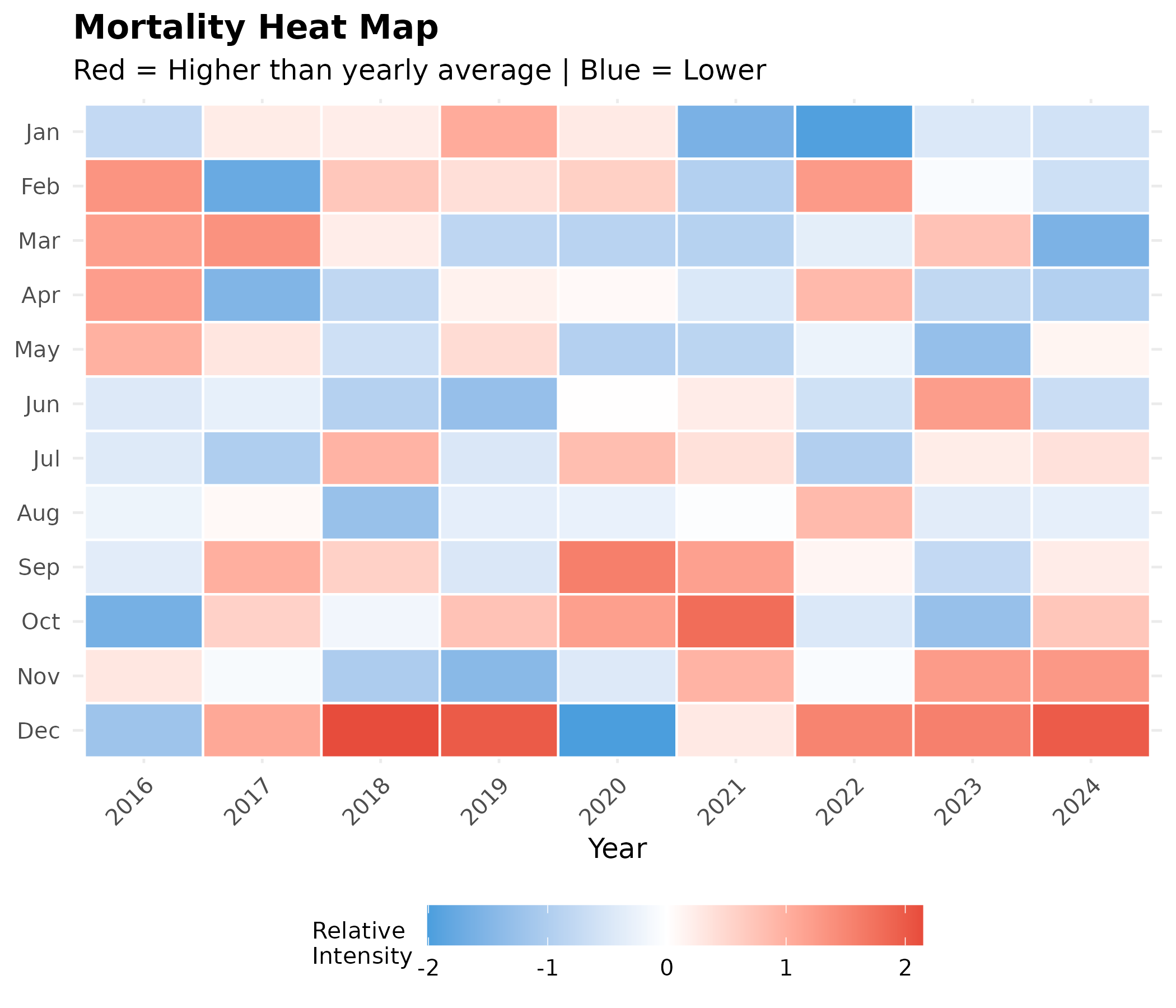
Interpretation:
- December Dominance: Contrary to the “entire winter peak” assumption, the heatmap shows that December (bottom row) is the only month consistently in the “Red Zone” (high mortality relative to each year’s average). January and February (top rows) are largely neutral (white) or even low (blue), reinforcing the finding that mortality intensity drops after the New Year.
- The 2021 Delta Shock: The “COVID Column” (2021) was not uniformly deadly. The first half of the year remained near average; the massive excess mortality occurred specifically in September, October, and November, visible as the dark red block in the lower portion of that column—coinciding with the Delta wave.
3. Statistical Confirmation
Seasonal Decomposition (STL)
To rigorously separate the trend, seasonality, and noise in mortality data, we use STL (Seasonal and Trend decomposition using Loess). This method is robust to outliers like the COVID-19 surge and provides a cleaner view of the underlying seasonal pattern.
# Convert to time series object using daily averages (normalized)
ts_deaths <- ts(deaths_monthly$daily_avg, start = c(2015, 1), frequency = 12)
# Perform STL decomposition
# s.window = "periodic" assumes seasonality is constant across years
decomp <- stl(ts_deaths, s.window = "periodic", robust = TRUE)
# Extract components for plotting
decomp_df <- data.frame(
date = deaths_monthly$date,
observed = as.numeric(ts_deaths),
trend = as.numeric(decomp$time.series[, "trend"]),
seasonal = as.numeric(decomp$time.series[, "seasonal"]),
remainder = as.numeric(decomp$time.series[, "remainder"])
) |>
tidyr::pivot_longer(
cols = c(observed, trend, seasonal, remainder),
names_to = "component",
values_to = "value"
) |>
mutate(
component = factor(component,
levels = c("observed", "trend", "seasonal", "remainder"),
labels = c("Observed (Daily Avg)", "Trend", "Seasonal Pattern", "Remainder (Noise)"))
)
# Plot decomposition
p <- decomp_df |>
ggplot(aes(x = date, y = value)) +
geom_line(color = "#2c3e50", linewidth = 0.6) +
geom_hline(yintercept = 0, linetype = "dashed", color = "grey60", linewidth = 0.3) +
facet_wrap(~component, ncol = 1, scales = "free_y") +
scale_x_date(date_breaks = "2 years", date_labels = "%Y") +
scale_y_continuous(labels = scales::comma) +
labs(
title = "STL Decomposition of Daily Average Mortality",
subtitle = "Separating trend, seasonality, and COVID-19 anomalies",
x = NULL,
y = "Deaths",
caption = "Note: COVID-19 surge appears in both Trend and Remainder components"
) +
theme_minimal(base_size = 12) +
theme(
plot.title = element_text(face = "bold"),
strip.text = element_text(face = "bold", size = 11),
panel.spacing = unit(0.5, "lines")
)
print(p)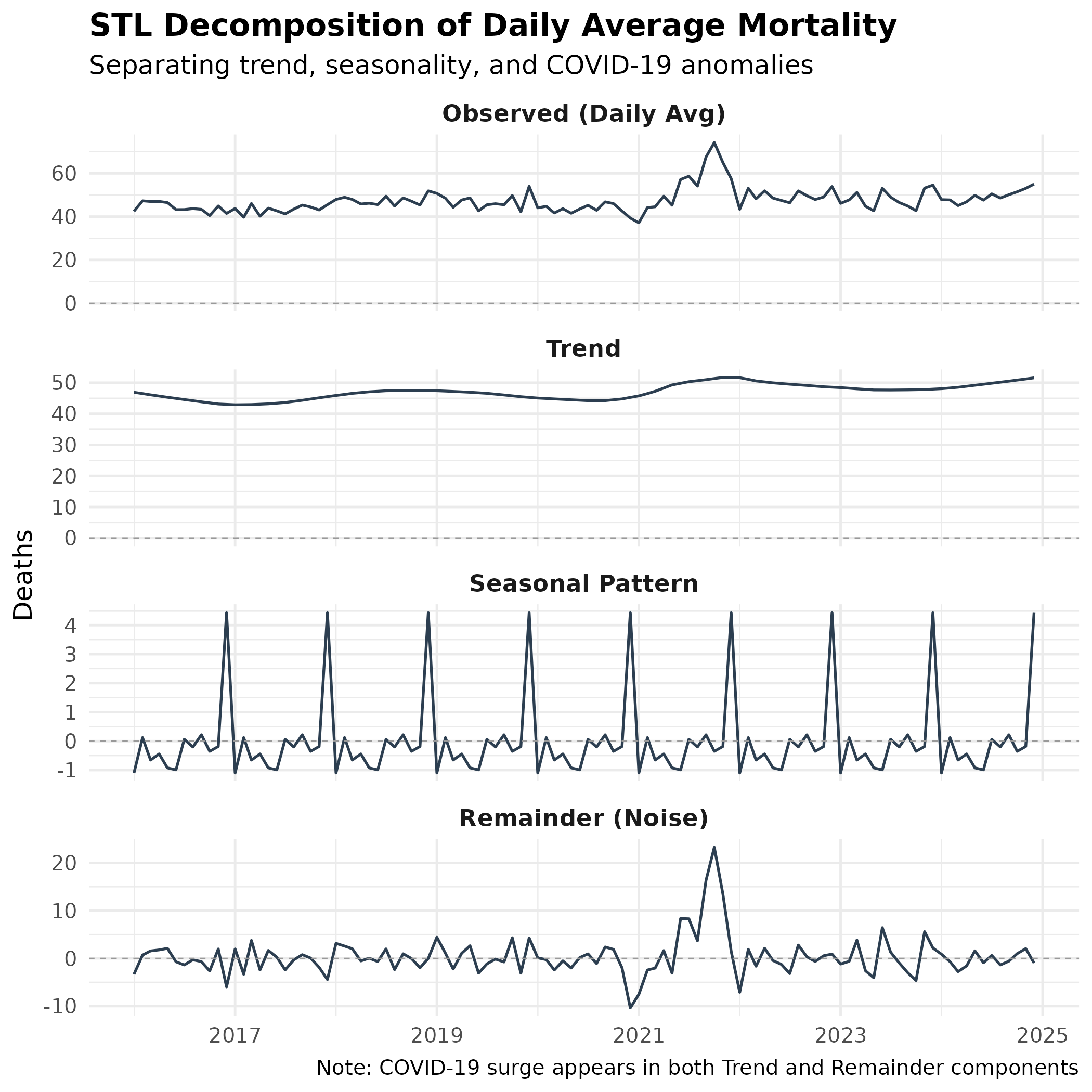
Interpretation:
- Trend: Shows the overall long-term pattern. Notice the sharp spike in 2021 (COVID-19 Delta wave) and gradual increase over time (population growth).
- Seasonal Pattern: A clean, repeating annual cycle emerges. Deaths peak in winter (Jan-Feb) and trough in summer (Jun-Aug). This pattern is consistent across all years.
- Remainder: Random noise plus anomalies. The large spikes in 2021 represent COVID-19 mortality that exceeded even the elevated trend.
Seasonal Pattern Analysis
Let’s examine the extracted seasonal component more closely:
# Extract one complete seasonal cycle (12 months)
seasonal_cycle <- data.frame(
month = 1:12,
month_name = factor(month.abb, levels = month.abb),
seasonal_effect = as.numeric(decomp$time.series[1:12, "seasonal"])
)
# Find peak and trough
peak_seasonal <- seasonal_cycle |> filter(seasonal_effect == max(seasonal_effect))
trough_seasonal <- seasonal_cycle |> filter(seasonal_effect == min(seasonal_effect))
# Seasonal pattern bar chart showing above/below average by month
p <- seasonal_cycle |>
ggplot(aes(x = month_name, y = seasonal_effect)) +
geom_col(aes(fill = ifelse(seasonal_effect > 0, "Above Average", "Below Average")),
alpha = 0.8) +
geom_hline(yintercept = 0, linetype = "solid", color = "grey40") + # zero line
geom_text(
aes(label = round(seasonal_effect, 1),
vjust = ifelse(seasonal_effect > 0, -0.5, 1.5)),
size = 3
) + # inline labels
scale_fill_manual(
values = c("Above Average" = "#e74c3c", "Below Average" = "#3498db"),
name = "Seasonal Effect"
) +
scale_y_continuous(labels = function(x) round(x, 1)) +
labs(
title = "Extracted Seasonal Pattern (Daily Average Deaths)",
subtitle = paste0("Peak: ", peak_seasonal$month_name, " (+", round(peak_seasonal$seasonal_effect, 1),
" deaths/day) | Trough: ", trough_seasonal$month_name, " (", round(trough_seasonal$seasonal_effect, 1), " deaths/day)"),
x = NULL,
y = "Daily deaths above/below average"
) +
theme_minimal(base_size = 12) +
theme(
plot.title = element_text(face = "bold"),
plot.subtitle = element_text(color = "grey40"),
legend.position = "bottom"
)
p # print static ggplot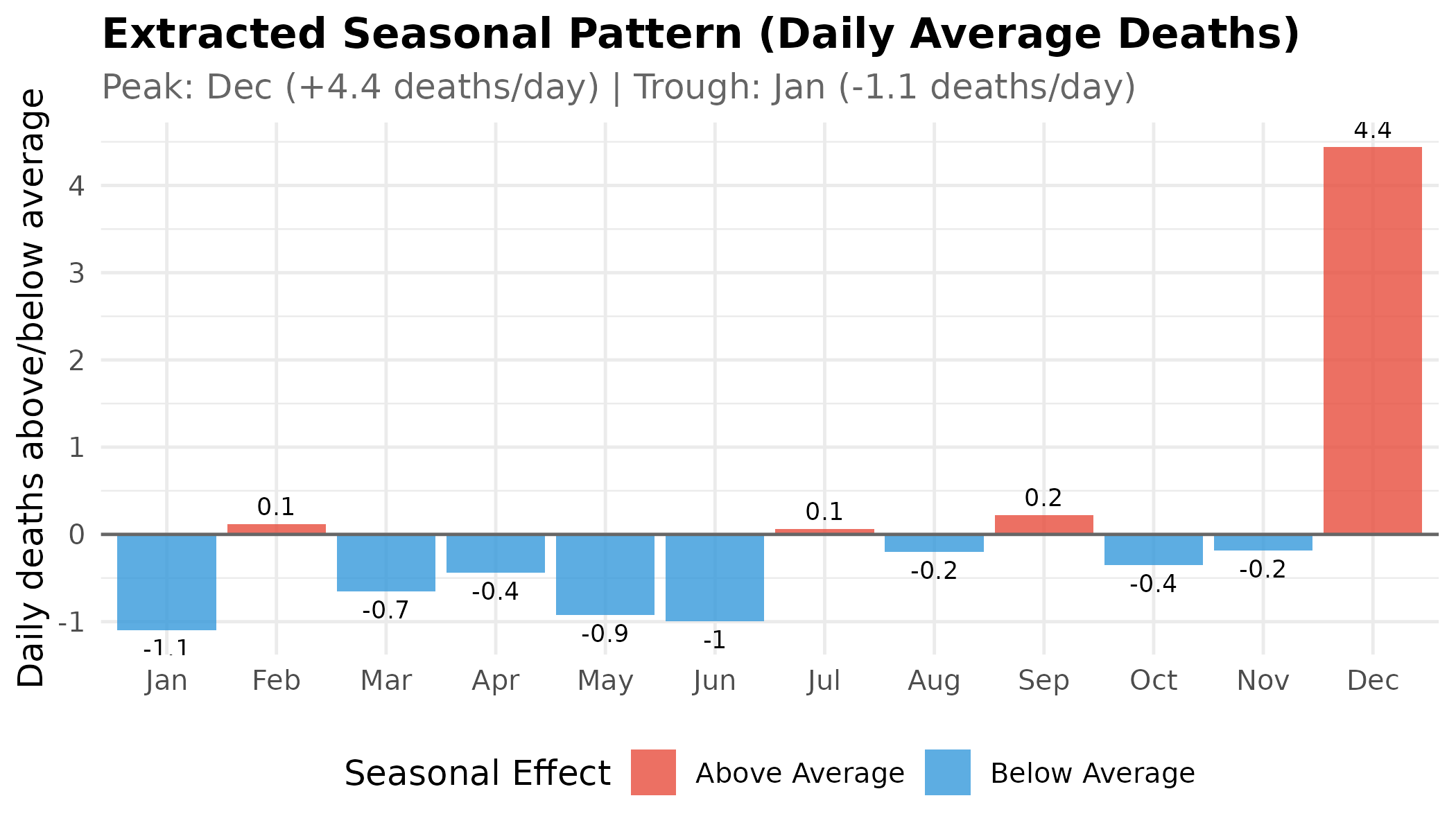
Key Finding: The STL decomposition confirms that Dec has the highest seasonal mortality, with approximately 4.4 more deaths per day than the annual average due to seasonality alone. The winter months (December–February) form a sustained high-mortality plateau.
Harmonic Regression Results
As a complementary approach, we use harmonic regression to mathematically model the seasonal cycle while controlling for the overall trend.
Caveat: Harmonic regression fits a symmetrical sine wave to the data. If the true seasonal pattern is asymmetric (e.g., a sharp winter spike with gradual decline), the model may shift the estimated peak to minimize overall residuals. Therefore, the STL decomposition above is more reliable for identifying exact peak timing.
# Fit harmonic regression with annual cycle (using daily averages)
deaths_monthly <- deaths_monthly |>
mutate(
time_index = as.numeric(date - min(date)) / 365.25,
sin_annual = sin(2 * pi * month / 12),
cos_annual = cos(2 * pi * month / 12)
)
# Fit model on daily averages
harmonic_model <- lm(daily_avg ~ time_index + sin_annual + cos_annual, data = deaths_monthly)
# Calculate amplitude and phase of seasonal component
coef_sin <- coef(harmonic_model)["sin_annual"]
coef_cos <- coef(harmonic_model)["cos_annual"]
amplitude <- sqrt(coef_sin^2 + coef_cos^2)
phase <- atan2(coef_sin, coef_cos) * 12 / (2 * pi)
peak_month_harmonic <- if (phase < 0) phase + 12 else phase
# Store results for display
harmonic_results <- list(
peak_month = month.name[round(peak_month_harmonic)],
amplitude = round(amplitude, 1),
r_squared = round(summary(harmonic_model)$r.squared * 100, 1)
)Statistical Findings
The harmonic regression model provides additional perspective on Mongolia’s mortality seasonality:
| Metric | Value |
|---|---|
| Estimated Peak Month | October |
| Seasonal Amplitude | ±1.4 deaths/day |
| Model Fit (R²) | 18.2% variance explained |
Methodological Note: The Harmonic Model estimates an October peak, which may contradict the observed December peak in the raw data. This is a model artifact. The harmonic method forces a symmetrical sine wave onto the data, whereas Mongolia’s mortality follows an asymmetrical “sharp spike” pattern in early winter. The model attempts to minimize overall error by “pulling” the estimated peak earlier to average out the curve. The STL Decomposition above is the more reliable indicator of peak timing.
4. Cause-Specific Analysis
To understand why deaths peak in winter, we need to examine the causes.
Note on Data Availability: Monthly mortality data by cause is not currently available in the public NSO API. We therefore use: 1. Annual Data to see long-term trends and the relative burden of each cause. 2. Monthly Case Data for respiratory diseases as a proxy for seasonal infection pressure.
Annual Trends by Cause
# Fetch deaths by cause (annual data)
cause_tbl <- "DT_NSO_2100_027V1"
indicators <- nso_dim_values(cause_tbl, "Indicator", labels = "en")
years <- nso_dim_values(cause_tbl, "Year", labels = "en")
cause_deaths <- nso_data(
tbl_id = cause_tbl,
selections = list(
"Indicator" = indicators$code,
"Region" = "0",
"Year" = years$code
),
labels = "en"
) |>
filter(!is.na(value)) |>
mutate(
year = as.integer(Year_en),
# Simplify cause names
Cause = case_when(
grepl("Injury", Indicator_en) ~ "Injuries & Poisoning",
grepl("cardiovascular", Indicator_en) ~ "Cardiovascular",
grepl("respiratory", Indicator_en) ~ "Respiratory",
grepl("digestive", Indicator_en) ~ "Digestive",
grepl("Cancer", Indicator_en) ~ "Cancer",
TRUE ~ Indicator_en
)
) |>
filter(Cause != "Deaths") # Exclude total
# Multi-line trend plot by cause of death
p <- cause_deaths |>
filter(year >= 2015) |>
ggplot(aes(x = year, y = value, color = Cause)) +
geom_line(linewidth = 1) +
geom_point(size = 2) +
scale_color_brewer(palette = "Set1") + # distinct colors for each cause
scale_y_continuous(labels = scales::comma) +
scale_x_continuous(breaks = 2015:2024) +
labs(
title = "Annual Deaths by Leading Cause (2015-2024)",
subtitle = "Cardiovascular disease is consistently the highest killer",
x = NULL,
y = "Number of Deaths per Year"
) +
theme_minimal(base_size = 12) +
theme(
plot.title = element_text(face = "bold"),
legend.position = "right"
)
p # print static ggplot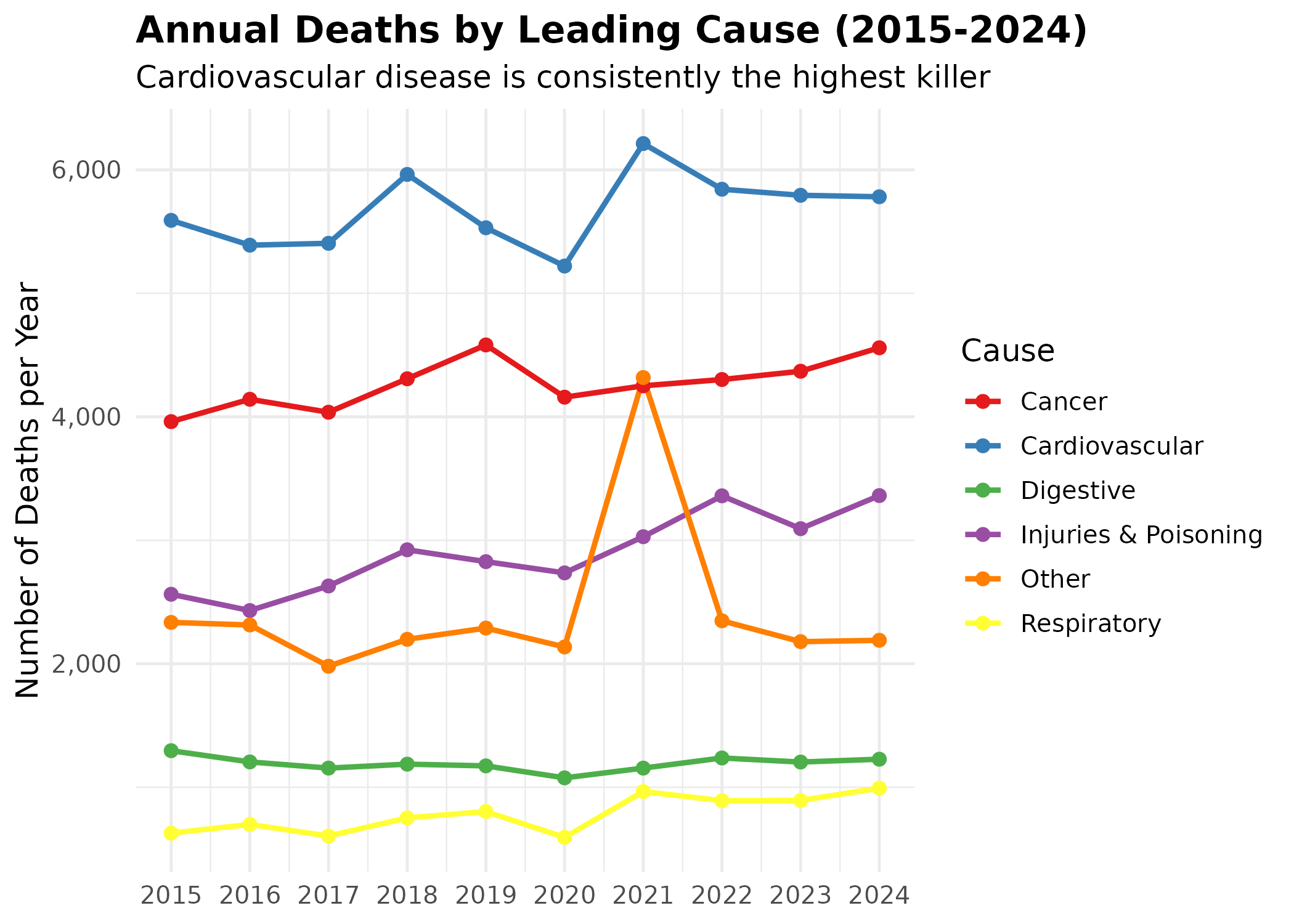
Interpretation:
- Cardiovascular Disease (Blue Line): This is the undisputed leading cause of death, hovering around 6,000 deaths annually. Its dominance suggests that any seasonal spike in total deaths is likely driven by cardiovascular stress from extreme cold.
- Cancer (Red Line): Consistently the second leading cause, showing a slow, steady upward trend over the decade.
- Injuries (Purple Line): While significant (3rd place), the trend is relatively flat. The lack of a massive winter spike in this category argues against the theory that New Year’s accidents are the primary driver of the December mortality peak.
- COVID-19 Impact: Note the sharp rise in “Respiratory” and other categories in 2021.
Respiratory Infections: A Seasonal Proxy
Since we cannot see monthly respiratory deaths, we look at monthly respiratory cases (e.g., flu, pneumonia) to see the seasonal pressure.
# Fetch monthly respiratory disease cases
comm_tbl <- "DT_NSO_2100_035V1"
# Code 12 is Respiratory infectious diseases
resp_cases <- nso_data(
tbl_id = comm_tbl,
selections = list(
"Indicators" = "12",
"Month" = months_filtered$code
),
labels = "en"
) |>
mutate(
date = as.Date(paste0(Month_en, "-01")),
month = month(date, label = TRUE)
) |>
filter(!is.na(value))
# Respiratory cases boxplot by month - proxy for seasonal infection pressure
p <- resp_cases |>
ggplot(aes(x = month, y = value)) +
geom_boxplot(fill = "#e74c3c", alpha = 0.6) +
scale_y_continuous(labels = scales::comma) +
labs(
title = "Seasonality of Respiratory Infections (Cases)",
subtitle = "Monthly distribution of reported cases (2015-2024)",
x = NULL,
y = "Reported Cases"
) +
theme_minimal(base_size = 12) +
theme(plot.title = element_text(face = "bold"))
p # print static ggplot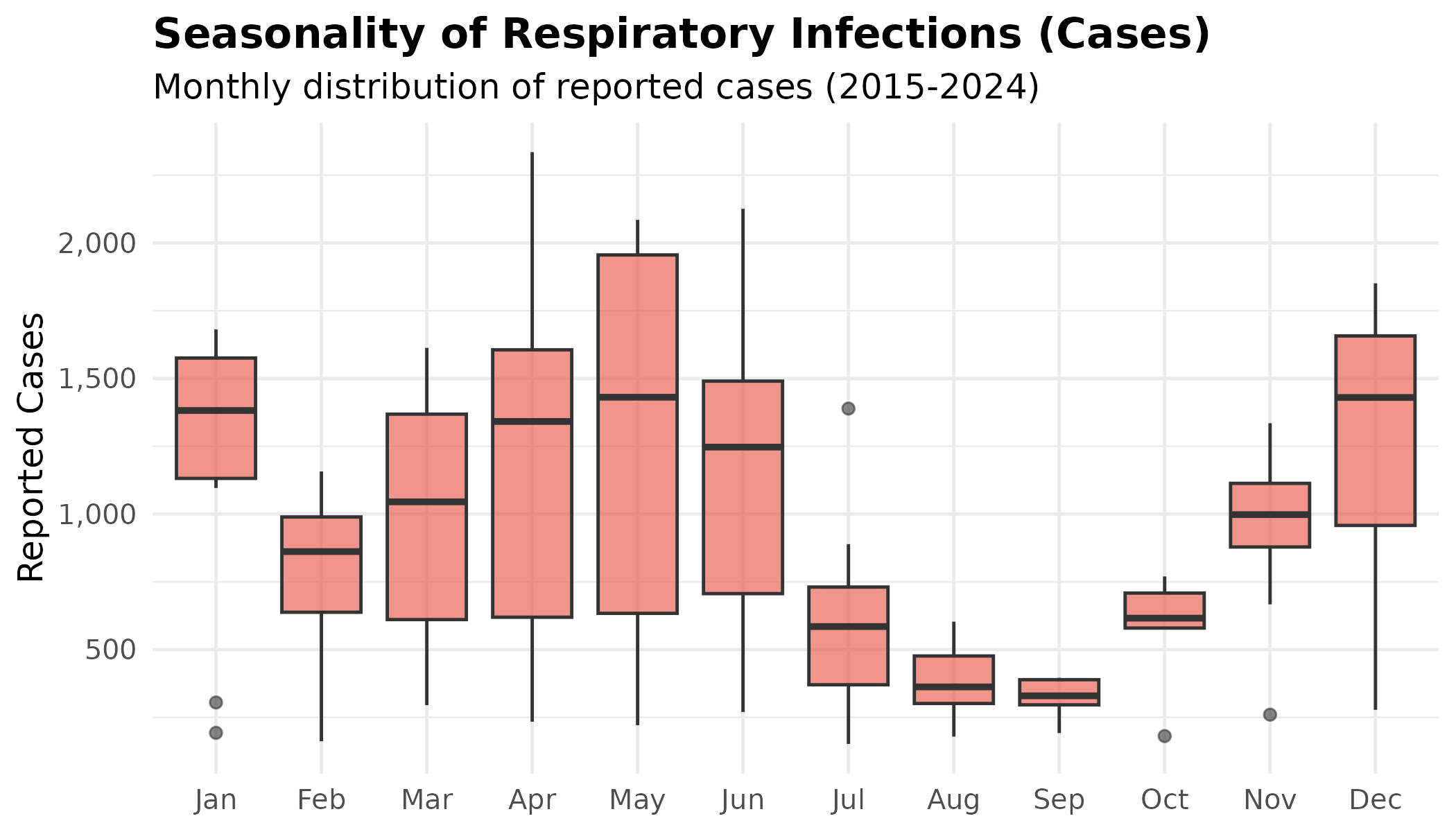
Interpretation:
- Complex Seasonality: Respiratory infections do not follow a simple winter curve. We see high case counts in December and January, followed by a notable dip in February, and a resurgence in May (spring peak).
- The Mortality Mismatch: While the December respiratory spike correlates with the December mortality peak, the May respiratory spike does not trigger a corresponding mortality spike (deaths are average in May). This suggests that while respiratory illness contributes to winter deaths, it is the combination of infection plus extreme cold (Dec/Jan) that is lethal. Infection alone (in warmer May) appears less fatal.
5. Regional Comparison
# Fetch deaths by region
regions_meta <- nso_dim_values(death_tbl, "Region", labels = "en")
# Get all regions' monthly data
regional_deaths <- nso_data(
tbl_id = death_tbl,
selections = list(
"Region" = regions_meta$code,
"Month" = months_filtered$code
),
labels = "en"
) |>
mutate(
date = as.Date(paste0(Month_en, "-01")),
year = year(date),
month = month(date),
month_name = month(date, label = TRUE, abbr = TRUE)
) |>
filter(!is.na(value), year >= 2015, year <= 2024)
# Classify as UB vs Rural
regional_deaths <- regional_deaths |>
mutate(
area_type = case_when(
Region_en == "Whole country" ~ "National",
grepl("Ulaanbaatar", Region_en) ~ "Ulaanbaatar",
TRUE ~ "Rural (Aimags)"
)
) |>
filter(area_type != "National")
# Aggregate by area type and month
area_monthly <- regional_deaths |>
group_by(area_type, month, month_name) |>
summarise(
mean_deaths = mean(value, na.rm = TRUE),
.groups = "drop"
)
# Normalize to percentage of annual mean for comparison
area_monthly <- area_monthly |>
group_by(area_type) |>
mutate(
annual_mean = mean(mean_deaths),
pct_of_mean = (mean_deaths / annual_mean - 1) * 100
) |>
ungroup()
# Urban vs Rural seasonality comparison
p <- area_monthly |>
mutate(month_name = factor(month_name, levels = month.abb)) |>
ggplot(aes(x = month_name, y = pct_of_mean, color = area_type, group = area_type)) +
geom_hline(yintercept = 0, linetype = "dashed", color = "grey50") + # baseline reference
geom_line(linewidth = 1.2) +
geom_point(size = 3) +
geom_text(aes(label = round(pct_of_mean, 0)), vjust = -1, size = 2.5) + # inline labels
scale_color_manual(
values = c("Ulaanbaatar" = "#e74c3c", "Rural (Aimags)" = "#3498db"),
name = NULL
) +
scale_y_continuous(expand = expansion(mult = c(0.1, 0.2))) +
labs(
title = "Urban vs Rural Seasonality",
subtitle = "Both follow the same winter pattern",
x = NULL,
y = "% Deviation from Annual Average"
) +
theme_minimal(base_size = 12) +
theme(
plot.title = element_text(face = "bold"),
legend.position = "top"
)
p # print static ggplot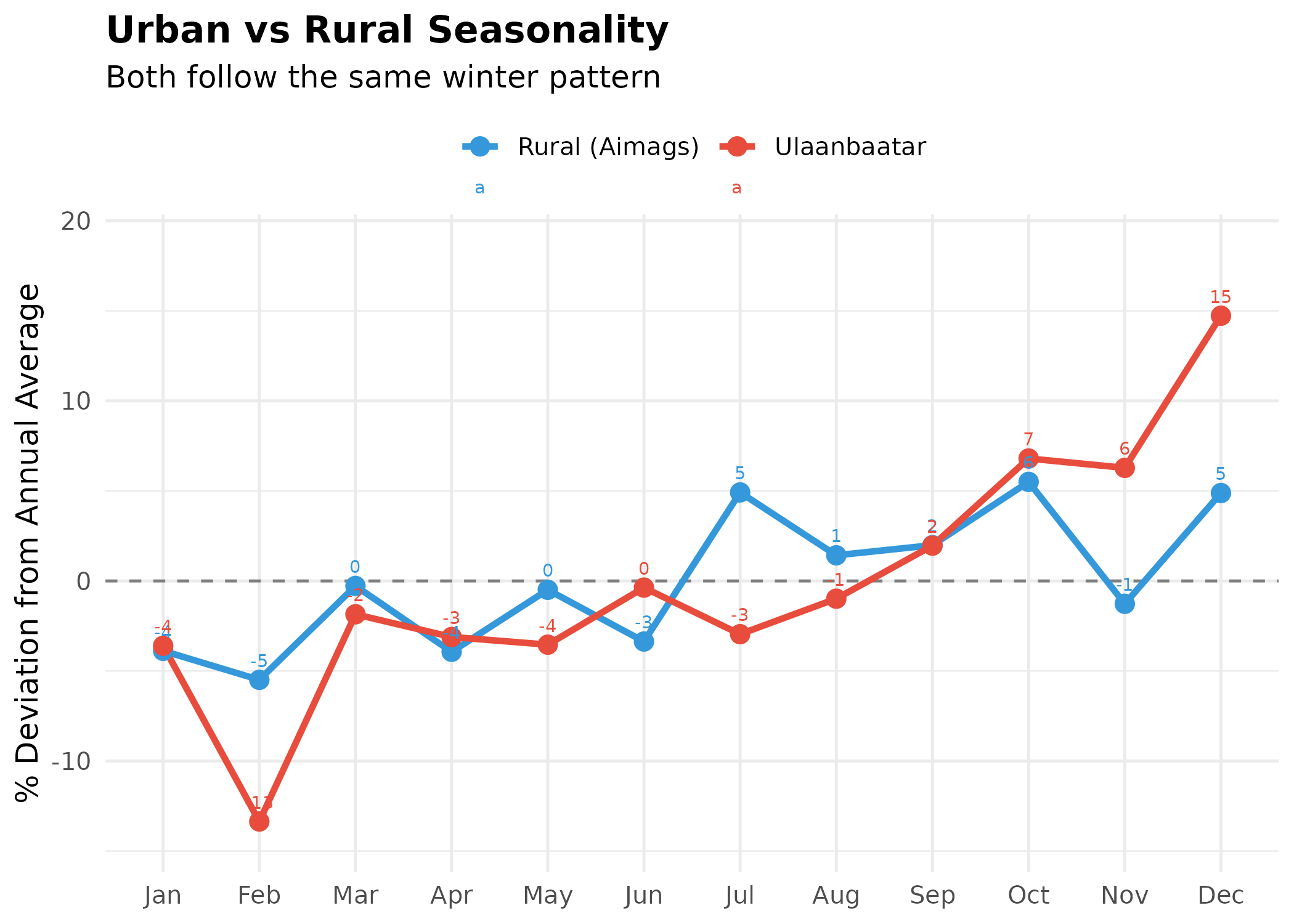
6. Hospital vs Total Deaths
Do hospital deaths show a different pattern? This can reveal healthcare access issues.
# Fetch hospital deaths
hospital_tbl <- "DT_NSO_2100_027V3"
hospital_months <- nso_dim_values(hospital_tbl, "Month", labels = "en")
# Filter to 2015-2024
hospital_months_filtered <- hospital_months |>
filter(grepl("^(2015|2016|2017|2018|2019|2020|2021|2022|2023|2024)-", label_en))
hospital_deaths <- nso_data(
tbl_id = hospital_tbl,
selections = list(
"Region" = "0",
"Month" = hospital_months_filtered$code
),
labels = "en"
) |>
mutate(
date = as.Date(paste0(Month_en, "-01")),
year = year(date),
month = month(date),
month_name = month(date, label = TRUE, abbr = TRUE),
type = "Hospital Deaths"
) |>
filter(!is.na(value), year >= 2015, year <= 2024)
# Combine with total deaths
comparison_data <- bind_rows(
deaths_monthly |> mutate(type = "All Deaths") |> select(date, year, month, month_name, value, type),
hospital_deaths |> select(date, year, month, month_name, value, type)
)
# Calculate monthly averages by type
type_monthly <- comparison_data |>
group_by(type, month, month_name) |>
summarise(mean_deaths = mean(value, na.rm = TRUE), .groups = "drop") |>
group_by(type) |>
mutate(
annual_mean = mean(mean_deaths),
pct_of_mean = (mean_deaths / annual_mean - 1) * 100
) |>
ungroup()
# Hospital vs All Deaths seasonality comparison
p <- type_monthly |>
mutate(month_name = factor(month_name, levels = month.abb)) |>
ggplot(aes(x = month_name, y = pct_of_mean, color = type, group = type)) +
geom_hline(yintercept = 0, linetype = "dashed", color = "grey50") + # baseline
geom_line(linewidth = 1.2) +
geom_point(size = 3) +
geom_text(aes(label = round(pct_of_mean, 0)), vjust = -1, size = 2.5) + # inline labels
scale_color_manual(
values = c("All Deaths" = "#2c3e50", "Hospital Deaths" = "#e74c3c"),
name = NULL
) +
scale_y_continuous(expand = expansion(mult = c(0.1, 0.2))) +
labs(
title = "Hospital vs All Deaths Seasonality",
subtitle = "Percentage deviation from annual mean",
x = NULL,
y = "% Deviation from Annual Average"
) +
theme_minimal(base_size = 12) +
theme(
plot.title = element_text(face = "bold"),
legend.position = "top"
)
p # print static ggplot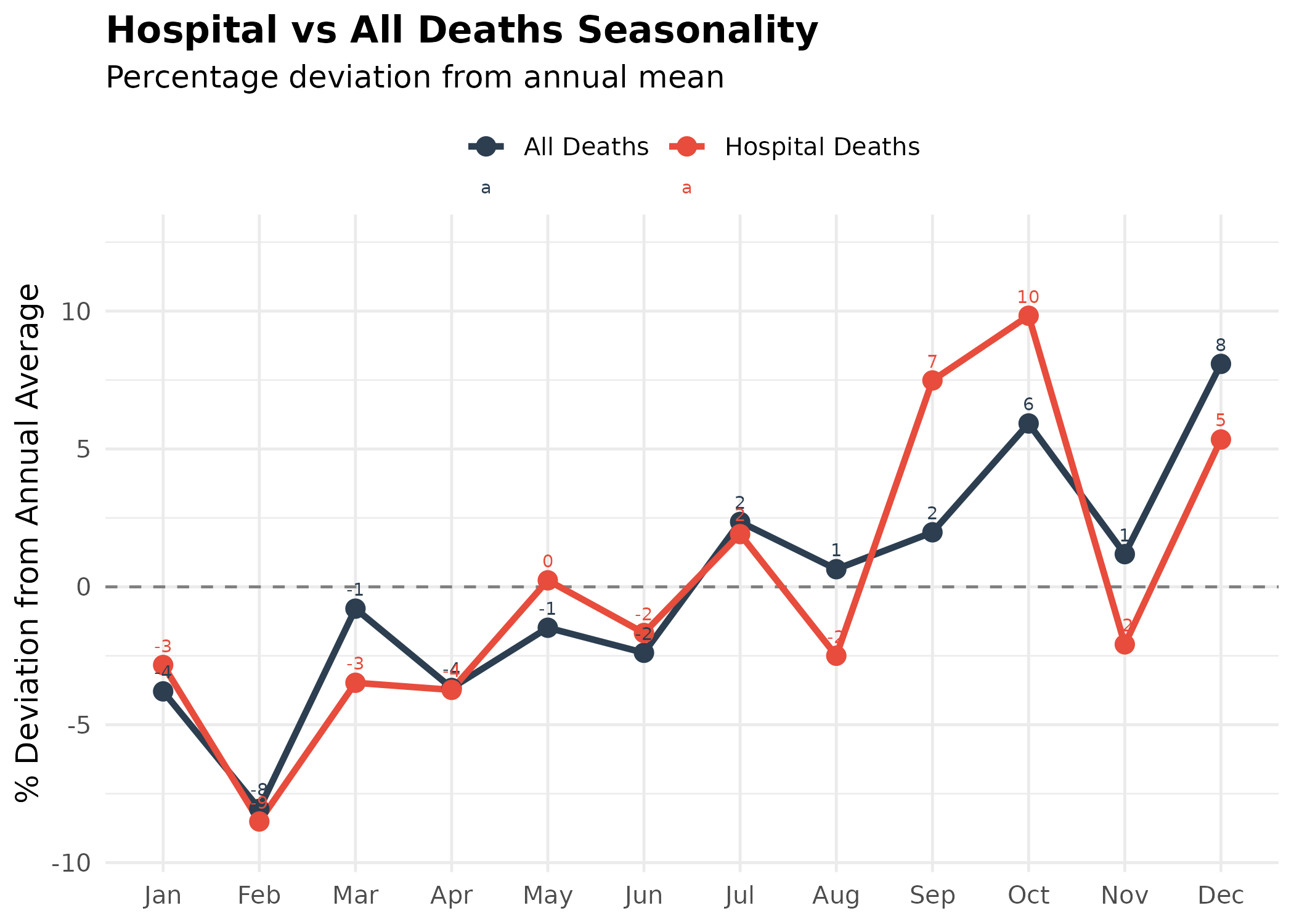
7. Detailed Summary Statistics
# Calculate summary statistics using daily averages
peak_analysis <- monthly_stats |>
mutate(
rank = rank(-mean_daily),
is_winter = month %in% c(11, 12, 1, 2, 3)
)
winter_avg <- peak_analysis |> filter(is_winter) |> summarise(mean = mean(mean_daily)) |> pull()
summer_avg <- peak_analysis |> filter(!is_winter) |> summarise(mean = mean(mean_daily)) |> pull()
winter_excess <- ((winter_avg / summer_avg) - 1) * 100
# Store results for inline display
summary_results <- list(
overall_avg = round(mean(monthly_stats$mean_daily), 1),
peak_month_name = as.character(peak_month$month_name),
peak_deaths = round(peak_month$mean_daily, 1),
trough_month_name = as.character(trough_month$month_name),
trough_deaths = round(trough_month$mean_daily, 1),
winter_excess = round(winter_excess, 1)
)8. Conclusions
Based on the evidence, we can address the social media claim:
-
Does December have the highest deaths?
- Yes, the data supports this. Unlike raw counts (where January’s 31 days inflate its ranking), the Daily Average analysis confirms that mortality intensity peaks in December (50.4 deaths/day). The social media claim is statistically accurate regarding the timing.
-
The Winter “Shock-and-Drop” Pattern:
- We observe a surprising pattern: mortality spikes sharply in December as temperatures plummet, followed by a dip in January (44.8 deaths/day—often the annual trough in daily averages). This contradicts the assumption that the entire winter is a sustained plateau; rather, the onset of extreme winter appears to be the deadliest phase. This is consistent with a “harvesting effect” where December’s cold shock claims the most vulnerable individuals first.
-
Is it because of New Year parties?
- We cannot determine this definitively. The NSO public API only provides annual death counts by cause—not monthly breakdowns. Without monthly cause-of-death data, we cannot confirm whether the December spike is driven by cardiovascular stress, respiratory illness, injuries, or some combination. While injuries appear to be a smaller fraction of annual deaths compared to cardiovascular disease, we lack the seasonal granularity to rule out a significant holiday-related injury spike in December specifically.
-
The COVID-19 Effect:
- The pandemic disrupted natural patterns. Lockdowns in 2020 actually suppressed mortality, while the 2021 waves caused historic highs. This highlights that infectious diseases (and our response to them) are major drivers of death rates.
Data Limitations
Important: This analysis is constrained by data availability. The NSO public API provides: - ✅ Monthly total deaths (which we analyzed) - ✅ Annual deaths by cause (which shows cardiovascular dominance yearly) - ❌ Monthly deaths by cause (not available)
To definitively answer why December mortality peaks, researchers would need access to monthly cause-of-death data, which may require direct collaboration with NSO or access to restricted datasets.
Final Verdict: The social media claim that “the highest deaths are occurring in December” is supported by the data when properly normalized for days in month. However, the underlying cause remains uncertain without monthly cause-specific mortality data. The correlation with extreme cold onset is suggestive but not conclusive.
Appendix: Reproducibility
sessionInfo()
#> R version 4.5.2 (2025-10-31)
#> Platform: x86_64-pc-linux-gnu
#> Running under: Ubuntu 24.04.3 LTS
#>
#> Matrix products: default
#> BLAS: /usr/lib/x86_64-linux-gnu/openblas-pthread/libblas.so.3
#> LAPACK: /usr/lib/x86_64-linux-gnu/openblas-pthread/libopenblasp-r0.3.26.so; LAPACK version 3.12.0
#>
#> locale:
#> [1] LC_CTYPE=C.UTF-8 LC_NUMERIC=C LC_TIME=C.UTF-8
#> [4] LC_COLLATE=C.UTF-8 LC_MONETARY=C.UTF-8 LC_MESSAGES=C.UTF-8
#> [7] LC_PAPER=C.UTF-8 LC_NAME=C LC_ADDRESS=C
#> [10] LC_TELEPHONE=C LC_MEASUREMENT=C.UTF-8 LC_IDENTIFICATION=C
#>
#> time zone: UTC
#> tzcode source: system (glibc)
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] scales_1.4.0 lubridate_1.9.4 ggplot2_4.0.1 tidyr_1.3.2
#> [5] dplyr_1.1.4 mongolstats_0.1.1
#>
#> loaded via a namespace (and not attached):
#> [1] gtable_0.3.6 jsonlite_2.0.0 compiler_4.5.2 tidyselect_1.2.1
#> [5] jquerylib_0.1.4 systemfonts_1.3.1 textshaping_1.0.4 yaml_2.3.12
#> [9] fastmap_1.2.0 R6_2.6.1 labeling_0.4.3 generics_0.1.4
#> [13] curl_7.0.0 httr2_1.2.2 knitr_1.51 htmlwidgets_1.6.4
#> [17] tibble_3.3.1 desc_1.4.3 RColorBrewer_1.1-3 bslib_0.9.0
#> [21] pillar_1.11.1 rlang_1.1.7 cachem_1.1.0 xfun_0.56
#> [25] S7_0.2.1 fs_1.6.6 sass_0.4.10 otel_0.2.0
#> [29] timechange_0.3.0 cli_3.6.5 withr_3.0.2 pkgdown_2.2.0
#> [33] magrittr_2.0.4 digest_0.6.39 grid_4.5.2 rappdirs_0.3.4
#> [37] lifecycle_1.0.5 vctrs_0.7.1 evaluate_1.0.5 glue_1.8.0
#> [41] farver_2.1.2 ragg_1.5.0 rmarkdown_2.30 purrr_1.2.1
#> [45] tools_4.5.2 pkgconfig_2.0.3 htmltools_0.5.9